受中国农业大学张勤教授邀请,美国威斯康辛大学(University of Wisconsin)Daniel Gianola教授和Guilherme Rosa教授将于2017年10月18-22日在中国农业大学举办数量遗传学高级短训班,Rosa 教授主要讲授数量遗传学和混合线性模型基础知识,Gianola教授则介绍线性模型、贝叶斯方法、GWAS、基因组选择、基因互作、非参数回归等前沿理论知识。欢迎广大同行和研究生积极报名参加。


Daniel Gianola Guilherme Rosa
一、主办单位
中国畜牧兽医学会动物遗传育种学分会
二、承办单位
畜禽育种国家工程实验室
农业部动物遗传育种与繁殖重点实验室
畜禽良种产业技术创新战略联盟
三、培训时间与地点
培训时间:2017年10月16-17日报到;10月18日-22日培训。培训地点:中国农业大学西校区(圆明园西路2号)。
四、培训费用与人数
培训费用:1200元/人,可优先参加10月17日举行的“动物基因组选择技术与产业化应用国际论坛”,会议详情另行通知。
培训人数:为保证培训质量,培训班人数将控制在40人之内。每个单位报名人数不超过2人。报名时请附简历。具体学员由学术组审核决定。
五、报到与住宿安排
报到地点:中国农业大学动物科技学院动科动医大楼。乘车路线请见附件四。
住宿地点:住宿自行解决,中国农业大学西校区附近宾馆详见附件三。
培训班提供午餐和晚餐,学员统一安排在中国农业大学校内餐厅就餐。
六、联系方式
曹 荣,13810613649,278734791@qq.com
丁向东,15811104301,xding@cau.edu.cn
地址:北京市海淀区圆明园西路2号中国农业大学动物科技学院
邮编:100193
七、注意事项
请申请培训人员务必于2017年9月31日前通过电子邮件发送回执至联系人信箱(邮件主题请注明“数量遗传学培训班回执”),以便妥善安排。回执请见下一页附件二。
中国畜牧兽医学会动物遗传育种学分会
2017年9月1日
附件一:培训安排
主讲教师 :
Professor Daniel Gianola(http://www.ansci.wisc.edu/Facultypages/gianola.html)Daniel Gianola 教授主要致力于数量遗传学理论研究,尤其对贝叶斯方法在动物育种中应用进行了开创性工作。Gianola教授著有多部学术论著,发表学术论文170余篇,担任多家学术杂志编委,多次在国际学术大会上做主题报告。根据2016年GOOGLE Scholar检索,Daniel Gianola 教授在基因组选择、贝叶斯方法、动物育种、动物遗传学领域论文引用排名分别位列第1,4,2和3名。鉴于其在数量遗传学领域的影响和贡献,Gianola教授获得了美国Iowa State University,丹麦奥胡斯,德国哥廷根大学等多所知名大学的荣誉教授。
Professor Guilherme Rosa(http://www.ansci.wisc.edu/Facultypages/rosa.html)Dr. Rosa is a Professor in quantitative genetics and genomics in the Departments of Animal Science and of Biostatistics & Medical Informatics at the UW-Madison. Before he joined the UW-Madison faculty, he was an Assistant Professor in the Department of Animal Sciences at Michigan State University (MSU), from January 2002 to April 2006. Dr. Rosa has a BS in Animal Science from the Sao Paulo State University (UNESP) in Brazil, and a MS in Animal Breeding from the same institution. He has also a PhD degree in Biostatistics from the University of Sao Paulo (USP), Brazil, and a post-doctoral training in statistical genetics at UW-Madison. His research program is at the interface between statistical/theoretical and molecular genetics and genomics, focusing on applications to animal models in domestic/managed and natural populations. His current projects include gene network and causal inference, optimal design of high throughput genomics assays, statistical modeling of high dimensional biological data, and integration of genomic information into genetic evaluation of farm animals. Dr. Rosa has (co)authored over 180 peer-reviewed publications and 10 book chapters, and has presented a number of invited talks and short courses in different countries. Among his many honors and recognitions, he has been recently awarded with the ‘2016 Rockefeller Prentice Memorial Award in Animal Breeding and Genetics’ from the American Society of Animal Science, and the ‘2017 Excellence in International Activities Award’ from the UW-Madison.
培训时间:10月18-22日 上午8:30-12:00 下午13:30-17:00 培训教材:培训讲义在报到时统一发放。
培训内容:
Professor Daniel Gianola
1. Introduction. Molecular markers and prediction. Predictive inference. Cross-validation. Overview of some penalized methods.
2. Review of least-squares, maximum likelihood and best linear unbiased prediction.
3. GWAS (genome-wide association study) and pitfalls.
4. Review of Bayesian inference, MCMC and Bayesian regression. Bayesian predictive distributions.
5. Challenges from complexity: over-parameterization, instrumental models, errors in gene action specification.
6. Genomic BLUP and genomic studentized prediction(GSTUP). The Bayesian alphabet (Bayes A B C, Bayesian Lasso, Bayes R)
7. The problem of dealing with gene-gene-gene-…-gene interactions.
8. Introduction to non-parametric regression: kernel methods and neural networks.
9. Estimating distributions of prediction errors via re-sampling.
Professor Guilherme Rosa
1.Quantitative genetics
Quantitative Genetics is the analysis of complex characters where both genetic and environment factors contribute to trait variation. Since this includes most traits of interest, such as disease susceptibility, crop yield, growth and reproduction in animals, human and animal behavior, and all gene expression data (transcriptome and proteome), a working knowledge of quantitative genetics is critical in diverse fields from plant and animal breeding, human genetics, genomics, behavior, to ecology and evolutionary biology. The course will cover the basics of quantitative genetics including: genetic basis for complex traits, population genetic assumptions including detection of admixture, Fisher’s variance decomposition, covariance between relatives, calculation of the numerator relationship matrix based on IBD alleles and an arbitrary pedigree, the genomic relationship matrix based on AIS alleles, heritability in the broad and narrow sense, inbreeding and crossbreeding, and response to selection. Also an introduction to advanced topics such as: Mixed Models, Best Linear Unbiased Prediction (BLUP), Genomic selection (GBLUP), Genome Wide Association Analysis (GWAS), QTL mapping, detection of selection from genomic data, correlated characters; and the multivariate response to selection.
2.Mixed Models in Quantitative Genetics
"Mixed models” refers to the analysis of linear models with arbitrary (co)variance structures among and within random effects and may be due to such factors as relationships or shared environments, cytoplasm, maternal effects and history. Mixed models are utilized in complex data analysis where the usual assumption(s) of independence and/or homogenous variances fail. Mixed models allow effects of nature to be separated from those of nurture and are emerging as the default method of analysis for human data. These issues are pervasive in human studies due to the lack of ability to randomize subjects to households, choice, and prior history. In plant breeding, growth and yield data are correlated due to shared locations, but diminish by distance resulting in spatial correlations. In animal breeding, performance data is correlated because individuals maybe related and may share common material environment as well as common pens or cages. Further, when individuals share a common space, they may experience indirect genetics effects (IGEs), which is an inherited effect in one individual experienced as an environmental effect in an associated individual. The evolution of cooperation and competition is based on IGEs, the estimation of which require mixed model analysis. Detection of cytoplasmic and epigenetic effects rely heavily on mixed model methods because of shared material or parental histories. Topics to be discussed include a basic matrix algebra review, the general linear model, derivation of the mixed model, BLUP and REML estimation, estimation and design issues, Bayesian formulations. Applications to be discussed include estimation of breeding values and genetic variances in general pedigrees, association mapping, genomic selection, spatial correlations and corrections, maternal genetic effects, detecting selection from genomic data, admixture detection and correction, direct and indirect genetic effects, models of general group and kin selection, genotype by environment interaction models.
附件二:培训班回执
数量遗传学培训班回执
姓 名 |
| 性别 |
| 年龄 |
|
类 别 | ¨教师 ¨博士生 ¨硕士生 |
参加10月17日“动物基因组选择国际论坛” ¨是 ¨否 |
单位名称 |
|
通讯地址(邮编) |
|
联系电话(手机) |
|
E-mail |
|
附件三: 中国农业大学西校区附近宾馆(由近到远)
北京农军宾馆(兰园小区东南)参考价:¥188 电话:(010)62899688 地址:竹园西街8号
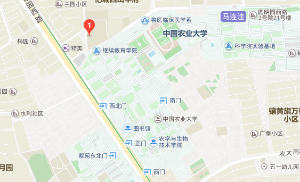
2.怡城林快捷酒店 参考价: ¥265 电话:(010)62896678 地址:圆明园西路9-5号,农大西校区路西,甜鸭梨南侧

3.英杰城宾馆 参考价: ¥168 电话:(010)62813378 地址:北京圆明园西路1栋2层
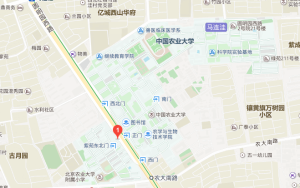
4.7天连锁酒店(北京农大南路店) 参考价: ¥379 电话:(010)62895767 地址:天秀路(309医院对面)
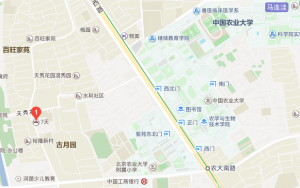
5.北京颐和园和颐酒店 参考价: ¥465 电话:(010)62828881 地址:北京海淀区天秀路9号 ,与龙背村路交叉口

6.汉庭酒店(北京颐和园店) 参考价: ¥351 电话:(010)62880909 地址:海淀区龙背村路101号(地铁4号线安河桥北B口出)

附件四:到中国农业大学西校区乘车路线
首都国际机场至中国农业大学西校区
线路1:机场大巴中关村线乘坐七站->中关村(保福寺桥)站->在保福寺桥西站换乘333内公交,乘坐10站->中国农业大学西校区站即到。
线路2:机场线乘坐二站->在三元桥站换乘地铁10号线,乘坐12站->苏州街站->在万泉桥南站乘坐651路公交,乘坐2站->中国农业大学西校区站即到。
线路3:出租车,约44公里,110-130元。
南苑机场至中国农业大学西校区
线路1:公交501路,乘坐14站->马家堡小区站->在马家堡站乘坐地铁4号线,乘坐21站-> 安河桥北站->骑共享单车约10分钟或乘坐575路公交,乘坐3站->中国农业大学西校区站即到。
线路2:南苑机场大巴公主坟专线乘坐8站->北京西站换乘特6公交,乘坐11站->中国农业大学西校区站即到。
线路3:出租车,约38公里,110-120元。
北京南站至中国农业大学西校区
线路1:地铁4号线,乘坐20站->安河桥北站->骑共享单车约10分钟或乘坐575路公交,乘坐3站->中国农业大学西校区站即到。
线路2:出租车,约28公里,约80-100元。
北京站至中国农业大学西校区
线路1:地铁2号线,乘坐9站->西直门换成地铁4号线->安河桥北站->骑共享单车约10分钟或乘坐575路公交,乘坐3站->中国农业大学西校区站即到。
线路2:出租车,约30公里,约90-100元。
北京西站至中国农业大学西校区
线路1:地铁4号线->安河桥北站->骑共享单车约10分钟或乘坐575路公交,乘坐3站->中国农业大学西校区站即到。
线路2:出租车,约20公里,约50-60元。